Numerical parameter identifiability¶
See the course by Marisa Eisenberg for an excellent introduction into the topic
In [1]:
Copied!
import numpy as np
from mxlpy import Model, Simulator, fns, plot
from mxlpy.identify import profile_likelihood
import numpy as np
from mxlpy import Model, Simulator, fns, plot
from mxlpy.identify import profile_likelihood
We start with an SIR model, which we use to generate some data (this would usually be experimentally measured data)
In [2]:
Copied!
def sir() -> Model:
return (
Model()
.add_variables({"s": 0.9, "i": 0.1, "r": 0.0})
.add_parameters({"beta": 0.2, "gamma": 0.1})
.add_reaction(
"infection",
fns.mass_action_2s,
args=["s", "i", "beta"],
stoichiometry={"s": -1, "i": 1},
)
.add_reaction(
"recovery",
fns.mass_action_1s,
args=["i", "gamma"],
stoichiometry={"i": -1, "r": 1},
)
)
data = Simulator(sir()).simulate(100).get_result().unwrap_or_err().variables
_ = plot.lines(data)
def sir() -> Model:
return (
Model()
.add_variables({"s": 0.9, "i": 0.1, "r": 0.0})
.add_parameters({"beta": 0.2, "gamma": 0.1})
.add_reaction(
"infection",
fns.mass_action_2s,
args=["s", "i", "beta"],
stoichiometry={"s": -1, "i": 1},
)
.add_reaction(
"recovery",
fns.mass_action_1s,
args=["i", "gamma"],
stoichiometry={"i": -1, "r": 1},
)
)
data = Simulator(sir()).simulate(100).get_result().unwrap_or_err().variables
_ = plot.lines(data)
We then, for n different values of each parameter we are interested in, we
- draw random samples for the remaining model parameters
- fit the model to the data (excluding the parameter we are interested in) and note the final error
- visualise the error for each parameter value
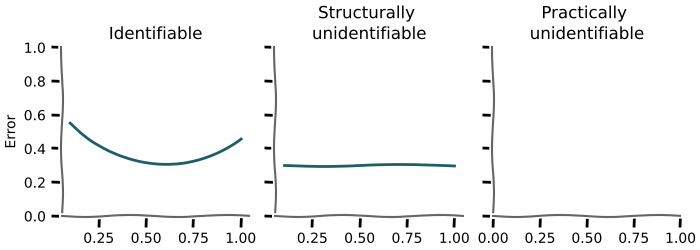
The error for a parameter should show a clear minimum around the different values used, otherwise it is not identifiable
In [3]:
Copied!
errors_beta = profile_likelihood(
sir(),
data=data,
parameter_name="beta",
parameter_values=np.linspace(0.2 * 0.5, 0.2 * 1.5, 10),
n_random=10,
)
fig, ax = plot.lines(errors_beta, legend=False)
ax.set(title="beta", xlabel="parameter value", ylabel="abs(error)")
plot.show()
errors_beta = profile_likelihood(
sir(),
data=data,
parameter_name="beta",
parameter_values=np.linspace(0.2 * 0.5, 0.2 * 1.5, 10),
n_random=10,
)
fig, ax = plot.lines(errors_beta, legend=False)
ax.set(title="beta", xlabel="parameter value", ylabel="abs(error)")
plot.show()
beta: 0%| | 0/10 [00:00<?, ?it/s]
beta: 10%|█ | 1/10 [00:01<00:13, 1.47s/it]
beta: 20%|██ | 2/10 [00:02<00:11, 1.46s/it]
beta: 30%|███ | 3/10 [00:04<00:10, 1.47s/it]
beta: 40%|████ | 4/10 [00:05<00:08, 1.46s/it]
beta: 50%|█████ | 5/10 [00:07<00:07, 1.50s/it]
beta: 60%|██████ | 6/10 [00:09<00:06, 1.55s/it]
beta: 70%|███████ | 7/10 [00:10<00:04, 1.55s/it]
beta: 80%|████████ | 8/10 [00:12<00:03, 1.52s/it]
beta: 90%|█████████ | 9/10 [00:13<00:01, 1.47s/it]
beta: 100%|██████████| 10/10 [00:14<00:00, 1.46s/it]
beta: 100%|██████████| 10/10 [00:14<00:00, 1.49s/it]