modelbase2¶
modelbase2 is a Python package designed to enable mechanistic learning, bridging the gap between mechanistic modeling and machine learning. The package enables you to integrate ordinary differential equation (ODE) models with data-driven techniques. This combination allows for more accurate and interpretable predictions in systems where both physical laws and data-driven insights are valuable. modelbase2 thus facilitates the development of models that are not only data-efficient but also robust and capable of capturing complex system dynamics. Choose one of the notebooks below to start your journey!
Building and simulating models¶
In this first notebook you will learn how to build ODE models and do basic simulations with them. This will allow you to create time courses and do steady-state analysis as shown below.
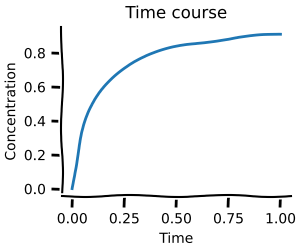
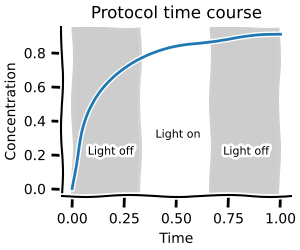
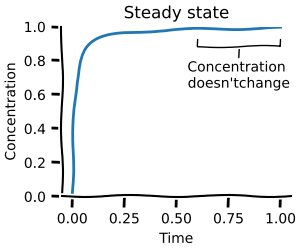
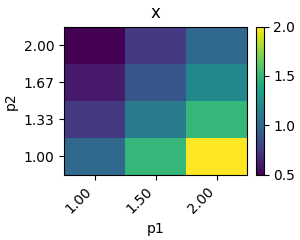
Parameter scans¶
Parameter scans allow you to systematically assess the behaviour of your model dependent on the value of one or more parameters. modelbase2 has rountines to scan over, and easily visualise time courses, protocol time courses, and steady states for one or more parameters.
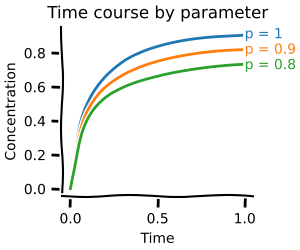
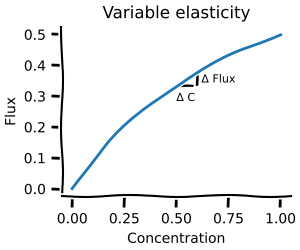
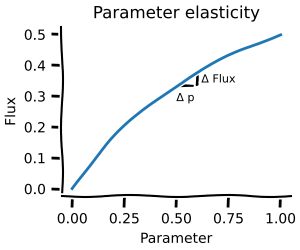
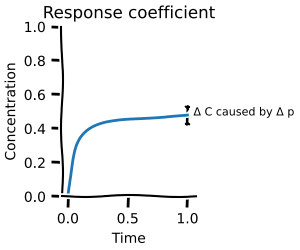
Metabolic control analysis¶
Metabolic control analysis answers the question: what happens to the concentrations and fluxes if I slightly perturb the system? It is thus a local measurement about which reactions hold the most control. If you ever read about rate-limiting steps, then this is for you!
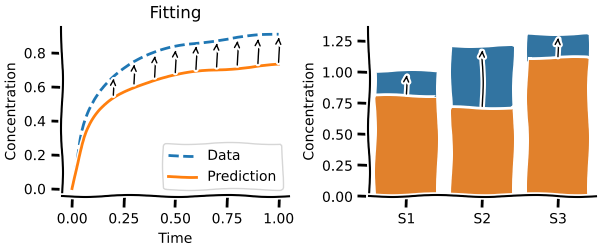
Fitting¶
Almost every model at some point needs to be fitted to experimental data to be validated. modelbase2 offers highly customisable routines for fitting either time series or steady-states.
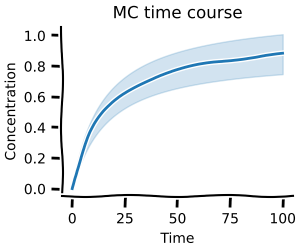
Monte Carlo methods¶
Almost every parameter in biology is better described with a distribution than a single value. Monte-carlo methods allow you to capture the range of possible behaviour your model can exhibit. This is especially useful when you want to understand the uncertainty in your model's predictions. modelbase2 offers these Monte Carlo methods for all scans ...
 +
+
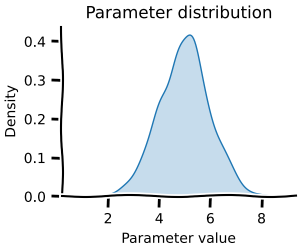 =
=
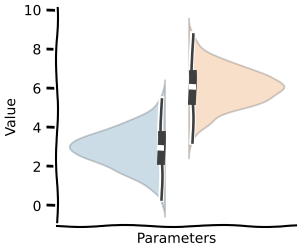
and even for metabolic control analysis
 +
+
 =
=
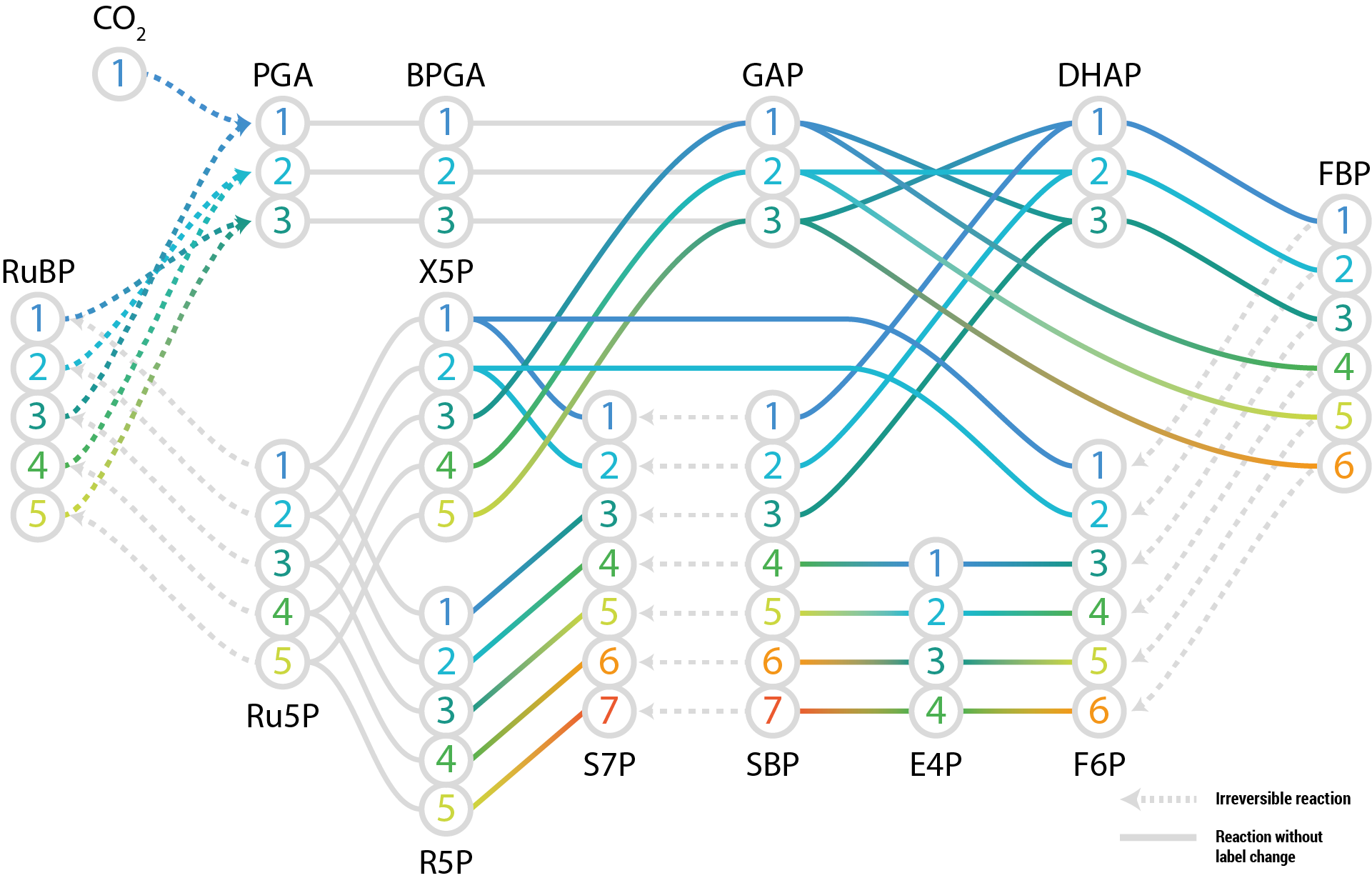
Label models¶
Labelled models allow explicitly mapping the transitions between isotopomers variables.
Mechanistic Learning¶
Mechanistic learning is the intersection of mechanistic modelling and machine learning. modelbase2 currently supports two such approaches: surrogates and neural posterior estimation. Surrogate models replace whole parts of a mechanistic model (or even the entire model) with machine learning models.
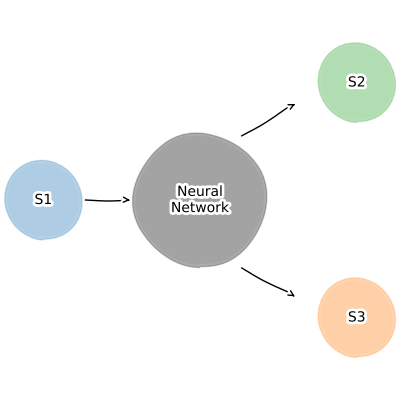
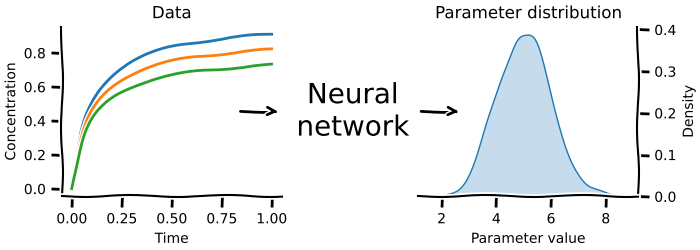
This allows combining together multiple models of arbitrary size, without having to worry about the internal state of each model. They are especially useful for improving the description of boundary effects, e.g. a dynamic description of downstream consumption. Neural posterior estimation answers the question: what parameters could have generated the data I measured? Here you use an ODE model and prior knowledge about the parameters of interest to create synthetic data. You then use the generated synthetic data as the features and the input parameters as the targets to train an inverse problem. Once that training is successful, the neural network can now predict the input parameters for real world data.
Parameterisation¶
Obtaining experimentally measured parameters can be challenging. Using the Brenda enzymes database we can obtain distributions of enzymatic parameters for a wide range of organisms. These distributions can in turn be used with our Monte-Carlo methods to capture the range of possible behaviour your model can exhibit.
 +
+
 =
=

Experimental features¶
A collection of experimental features for you to explore. Warning: all APIs shown should be considered unstable and may change without notice.