from pathlib import Path
import matplotlib.pyplot as plt
from modelbase2 import Model, fns, mc, plot
from modelbase2.distributions import GaussianKde, sample
from modelbase2.parameterise import get_km_and_kcat_from_brenda
Model parameterisation¶
Obtaining experimentally measured parameters can be challenging.
Using the Brenda enzymes database we can obtain distributions of enzymatic parameters for a wide range of organisms.
We can do that with the modelbase2.parameterise module.
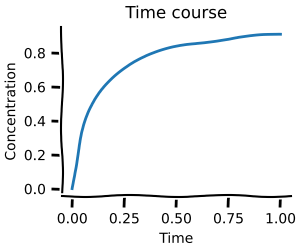
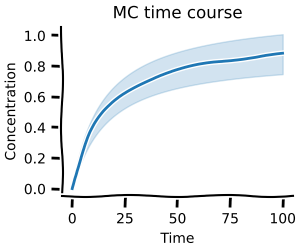
These distributions can then in turn be used with our Monte-Carlo methods to capture the range of possible behaviour your model can exhibit.
 +
+
 =
=
In order to obtain the parameters for a given Enzyme commision number (ec) we will manually download the database.
You have to do this manually due to the brenda licensing terms.
Note: we have created a small copy of just the rubisco data here to keep the documentation running.
Adjust yourbrenda_pathaccordingly
kms, kcats = get_km_and_kcat_from_brenda(
ec="4.1.1.39",
brenda_path=Path("assets") / "brenda_rubisco_only.json",
)
print(f"Found: {len(kms)} michaelis constants")
kms.head()
Found: 668 michaelis constants
| value | substrate | organism | uniprot | sequence | |
|---|---|---|---|---|---|
| 0 | 0.0290 | CO2 | Amphicarpaea bracteata | A0A1C3HPM0 | MSPQTETKASVGFKAGVKDYKLTYYTPDYETKDTDILAAFRVTPQP... |
| 5 | 0.0510 | CO2 | Archaeoglobus fulgidus | O28635 | MAEFEIYREYVDKSYEPQKDDIVAVFRITPAEGFTIEDAAGAVAAE... |
| 7 | 0.0279 | CO2 | Hordeum murinum | A0A1C3HPQ4 | MSPQTETKAGVGFKAGVKDYKLTYYTPEYETKDTDILAAFRVSPQP... |
| 8 | 0.0279 | CO2 | Hordeum brachyantherum | A0A1C3HPQ0 | MSPQTETKAGVGFQAGVKDYKLTYYTPEYETKDTDILAAFRVSPQP... |
| 9 | 0.0195 | CO2 | Glycine canescens | A0A1C3HPP9 | MSPQTETKASVGFKAGVKDYKLTYYTPDYETKDTDILAAFRVTPQP... |
As you can see above, this provides you with parameter values for different organisms and substrates.
Thus, we first filter by the specific substrate we are interested in.
# Filter out a specific substrate
kms = kms[kms["substrate"] == "CO2"]
kcats = kcats[kcats["substrate"] == "CO2"]
print(f"Filtered to {len(kms)} michaelis constants")
kms.head()
Filtered to 443 michaelis constants
| value | substrate | organism | uniprot | sequence | |
|---|---|---|---|---|---|
| 0 | 0.0290 | CO2 | Amphicarpaea bracteata | A0A1C3HPM0 | MSPQTETKASVGFKAGVKDYKLTYYTPDYETKDTDILAAFRVTPQP... |
| 5 | 0.0510 | CO2 | Archaeoglobus fulgidus | O28635 | MAEFEIYREYVDKSYEPQKDDIVAVFRITPAEGFTIEDAAGAVAAE... |
| 7 | 0.0279 | CO2 | Hordeum murinum | A0A1C3HPQ4 | MSPQTETKAGVGFKAGVKDYKLTYYTPEYETKDTDILAAFRVSPQP... |
| 8 | 0.0279 | CO2 | Hordeum brachyantherum | A0A1C3HPQ0 | MSPQTETKAGVGFQAGVKDYKLTYYTPEYETKDTDILAAFRVSPQP... |
| 9 | 0.0195 | CO2 | Glycine canescens | A0A1C3HPP9 | MSPQTETKASVGFKAGVKDYKLTYYTPDYETKDTDILAAFRVTPQP... |
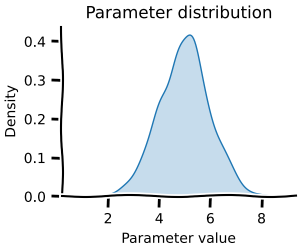
Since these are sufficiently many values, we can create a Gaussian Kernel Density estimate of them.
km_dist = GaussianKde.from_data(kms["value"])
fig, ax = km_dist.plot(
xmin=kms["value"].min() * 0.8,
xmax=kms["value"].max() * 1.2,
)
ax.set(title=f"rubisco km for CO2, n={len(kms)}")
plt.show()
This kernel density estimate we can now use exactly like other distribution in our Monte-Carlo routines (see the Monte Carlo notebook for more information).
Here, we create a small toy model and then use the distribution obtained from the experimental data to calculate the steady-state distribution of the model concentration.
model = (
Model()
.add_parameters({"k_out": 1.0, "km": 1.0})
.add_variable("PGA", 0)
.add_reaction(
"rubisco",
fns.constant,
args=["km"],
stoichiometry={"PGA": 2},
)
.add_reaction(
"outflux",
fns.mass_action_1s,
args=["PGA", "k_out"],
stoichiometry={"PGA": -1},
)
)
ss = mc.steady_state(model, mc_parameters=sample({"km": km_dist}, n=10))
fig, ax = plt.subplots(figsize=(4, 3))
ax.set(ylabel="Steady-state concentration")
plot.violins(ss.concs, ax=ax)
plt.show()
0%| | 0/10 [00:00<?, ?it/s]
100%|██████████| 10/10 [00:00<00:00, 67.41it/s]
First finish line
With that you now know most of what you will need from a day-to-day basis about parameter scans in modelbase2.Congratulations!