from __future__ import annotations
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import modelbase2 as mb2
from example_models import (
get_lin_chain_two_circles,
get_linear_chain_2v,
get_upper_glycolysis,
)
from modelbase2 import make_protocol, mc, mca, plot
Monte Carlo methods¶
Almost every parameter in biology is better described with a distribution than a single value.
Monte-carlo methods allow you to capture the range of possible behaviour your model can exhibit.
This is especially useful when you want to understand the uncertainty in your model's predictions.
modelbase2 offers these Monte Carlo methods for all scans ...
 +
+
 =
=
and even for metabolic control analysis
 +
+
 =
=
In this tutorial we will mostly use the modelbase2.distributions and modelbase2.mca modules, which contain the functionality to sample from distributions and run distributed analyses.
Sample values¶
To do any Monte-Carlo analysis, we first need to be able to sample values.
For that, you can use the sample function and distributions supplied by modelbase2.
These are mostly thin wrappers around the numpy and scipy sampling methods.
from modelbase2.distributions import LogNormal, Uniform, sample
sample(
{
"k2": Uniform(1.0, 2.0),
"k3": LogNormal(mean=1.0, sigma=1.0),
},
n=5,
)
| k2 | k3 | |
|---|---|---|
| 0 | 1.773956 | 0.739205 |
| 1 | 1.438878 | 3.088978 |
| 2 | 1.858598 | 1.981308 |
| 3 | 1.697368 | 2.672993 |
| 4 | 1.094177 | 1.158303 |
Steady-state¶
Using mc.steady_state you can calculate the steady-state distribution given the monte-carlo parameters.
This works analogously to the scan.steady_state function, except the index of the dataframes is always just an integer.
The parameters used can be obtained by result.parameters.
We will use a linear chain of reactions with two circles as an example model for this notebook.
$$ \begin{array}{c|c} \mathrm{Reaction} & \mathrm{Stoichiometry} \\ \hline v_0 & \varnothing \rightarrow{} \mathrm{x_1} \\ v_1 & -\mathrm{x_1} \rightarrow{} \mathrm{x_2} \\ v_2 & -\mathrm{x_1} \rightarrow{} \mathrm{x_3} \\ v_3 & -\mathrm{x_1} \rightarrow{} \mathrm{x_4} \\ v_4 & -\mathrm{x_4} \rightarrow{} \varnothing\\ v_5 & -\mathrm{x_2} \rightarrow{} \mathrm{x_1} \\ v_6 & -\mathrm{x_3} \rightarrow{} \mathrm{x_1} \\ \end{array} $$
ss = mc.steady_state(
get_linear_chain_2v(),
mc_parameters=sample(
{
"k1": Uniform(0.9, 1.1),
"k2": Uniform(1.0, 1.3),
"k3": LogNormal(mean=1.0, sigma=0.2),
},
n=10,
),
)
fig, (ax1, ax2) = plot.two_axes(figsize=(6, 2.5), sharex=False)
plot.violins(ss.concs, ax=ax1)
plot.violins(ss.fluxes, ax=ax2)
ax1.set(xlabel="Variables", ylabel="Concentration / a.u.")
ax2.set(xlabel="Reactions", ylabel="Flux / a.u.")
plt.show()
0%| | 0/10 [00:00<?, ?it/s]
50%|█████ | 5/10 [00:00<00:00, 49.31it/s]
100%|██████████| 10/10 [00:00<00:00, 66.94it/s]
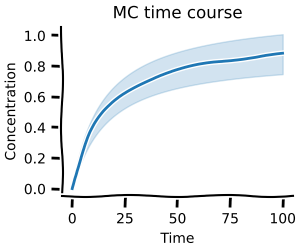
Time course¶
Using mc.time_course you can calculate time courses for sampled parameters.
 +
+
 =
=

This function works analogously to scan.time_course.
The pandas.DataFrames for concentrations and fluxes have a n x time pandas.MultiIndex.
The corresponding parameters can be found in result.parameters
tc = mc.time_course(
get_linear_chain_2v(),
time_points=np.linspace(0, 1, 11),
mc_parameters=sample(
{
"k1": Uniform(0.9, 1.1),
"k2": Uniform(1.0, 1.3),
"k3": LogNormal(mean=1.0, sigma=0.2),
},
n=10,
),
)
fig, (ax1, ax2) = plot.two_axes(figsize=(7, 4))
plot.lines_mean_std_from_2d_idx(tc.concs, ax=ax1)
plot.lines_mean_std_from_2d_idx(tc.fluxes, ax=ax2)
ax1.set(xlabel="Time / a.u", ylabel="Concentration / a.u.")
ax2.set(xlabel="Time / a.u", ylabel="Flux / a.u.")
plt.show()
0%| | 0/10 [00:00<?, ?it/s]
100%|██████████| 10/10 [00:00<00:00, 98.23it/s]
100%|██████████| 10/10 [00:00<00:00, 57.66it/s]
Protocol time course¶
Using mc.time_course_over_protocol you can calculate time courses for sampled parameters given a discrete protocol.
 +
+
 =
=
The pandas.DataFrames for concentrations and fluxes have a n x time pandas.MultiIndex.
The corresponding parameters can be found in scan.parameters
tc = mc.time_course_over_protocol(
get_linear_chain_2v(),
time_points_per_step=10,
protocol=make_protocol(
[
(1, {"k1": 1}),
(2, {"k1": 2}),
(3, {"k1": 1}),
]
),
mc_parameters=sample(
{
"k2": Uniform(1.0, 1.3),
"k3": LogNormal(mean=1.0, sigma=0.2),
},
n=10,
),
)
fig, (ax1, ax2) = plot.two_axes(figsize=(7, 4))
plot.lines_mean_std_from_2d_idx(tc.concs, ax=ax1)
plot.lines_mean_std_from_2d_idx(tc.fluxes, ax=ax2)
for ax in (ax1, ax2):
plot.shade_protocol(tc.protocol["k1"], ax=ax, alpha=0.1)
ax1.set(xlabel="Time / a.u", ylabel="Concentration / a.u.")
ax2.set(xlabel="Time / a.u", ylabel="Flux / a.u.")
plt.show()
0%| | 0/10 [00:00<?, ?it/s]
10%|█ | 1/10 [00:00<00:00, 9.84it/s]
100%|██████████| 10/10 [00:00<00:00, 36.50it/s]
Metabolic control analysis¶
modelbase2 further has routines for monte-carlo distributed metabolic control analysis.
This allows quantifying, whether the coefficients obtained from the MCA analysis are robust against parameter changes or whether they are just an artifact of a particular choice of parameters.
mc_elas = mc.variable_elasticities(
get_upper_glycolysis(),
concs={
"GLC": 0.3,
"G6P": 0.4,
"F6P": 0.5,
"FBP": 0.6,
"ATP": 0.4,
"ADP": 0.6,
},
variables=["GLC", "F6P"],
mc_parameters=sample(
{
# "k1": LogNormal(mean=np.log(0.25), sigma=1.0),
# "k2": LogNormal(mean=np.log(1.0), sigma=1.0),
"k3": LogNormal(mean=np.log(1.0), sigma=1.0),
# "k4": LogNormal(mean=np.log(1.0), sigma=1.0),
# "k5": LogNormal(mean=np.log(1.0), sigma=1.0),
# "k6": LogNormal(mean=np.log(1.0), sigma=1.0),
# "k7": LogNormal(mean=np.log(2.5), sigma=1.0),
},
n=5,
),
)
_ = plot.violins_from_2d_idx(mc_elas)
plt.show()
0%| | 0/5 [00:00<?, ?it/s]
100%|██████████| 5/5 [00:00<00:00, 34.13it/s]
Parameter elasticities¶
 +
+
 =
=

elas = mc.parameter_elasticities(
get_upper_glycolysis(),
concs={
"GLC": 0.3,
"G6P": 0.4,
"F6P": 0.5,
"FBP": 0.6,
"ATP": 0.4,
"ADP": 0.6,
},
parameters=["k1", "k2", "k3"],
mc_parameters=sample(
{
"k3": LogNormal(mean=np.log(0.25), sigma=1.0),
},
n=5,
),
)
_ = plot.violins_from_2d_idx(elas)
plt.show()
0%| | 0/5 [00:00<?, ?it/s]
100%|██████████| 5/5 [00:00<00:00, 33.34it/s]
Response coefficients¶
 +
+
 =
=

# Compare with "normal" control coefficients
rc = mca.response_coefficients(
get_lin_chain_two_circles(),
parameters=["vmax_1", "vmax_2", "vmax_3", "vmax_5", "vmax_6"],
)
_ = plot.heatmap(rc.concs)
mrc = mc.response_coefficients(
get_lin_chain_two_circles(),
parameters=["vmax_1", "vmax_2", "vmax_3", "vmax_5", "vmax_6"],
mc_parameters=sample(
{
"k0": LogNormal(np.log(1.0), 1.0),
"k4": LogNormal(np.log(0.5), 1.0),
},
n=10,
),
)
_ = plot.violins_from_2d_idx(mrc.concs, n_cols=len(mrc.concs.columns))
0%| | 0/5 [00:00<?, ?it/s]
20%|██ | 1/5 [00:00<00:01, 2.60it/s]
100%|██████████| 5/5 [00:00<00:00, 10.66it/s]
100%|██████████| 5/5 [00:00<00:00, 7.50it/s]
0%| | 0/10 [00:00<?, ?it/s]
0%| | 0/5 [00:00<?, ?it/s]
0%| | 0/5 [00:00<?, ?it/s]
0%| | 0/5 [00:00<?, ?it/s]
0%| | 0/5 [00:00<?, ?it/s]
0%| | 0/5 [00:00<?, ?it/s]
0%| | 0/5 [00:00<?, ?it/s]
20%|██ | 1/5 [00:00<00:01, 3.31it/s]
20%|██ | 1/5 [00:00<00:01, 2.33it/s]
20%|██ | 1/5 [00:00<00:01, 2.05it/s]
20%|██ | 1/5 [00:00<00:01, 2.40it/s]
40%|████ | 2/5 [00:00<00:00, 3.17it/s]
40%|████ | 2/5 [00:00<00:01, 2.30it/s]
60%|██████ | 3/5 [00:00<00:00, 3.20it/s]
40%|████ | 2/5 [00:00<00:01, 2.04it/s]
40%|████ | 2/5 [00:00<00:01, 2.34it/s]
80%|████████ | 4/5 [00:01<00:00, 3.22it/s]
60%|██████ | 3/5 [00:01<00:00, 2.32it/s]
60%|██████ | 3/5 [00:01<00:00, 2.35it/s]
60%|██████ | 3/5 [00:01<00:01, 2.00it/s]
100%|██████████| 5/5 [00:01<00:00, 3.19it/s]
100%|██████████| 5/5 [00:01<00:00, 3.18it/s]
10%|█ | 1/10 [00:01<00:14, 1.62s/it]
0%| | 0/5 [00:00<?, ?it/s]
80%|████████ | 4/5 [00:01<00:00, 2.34it/s]
0%| | 0/5 [00:00<?, ?it/s]
0%| | 0/5 [00:00<?, ?it/s]
80%|████████ | 4/5 [00:01<00:00, 2.39it/s]
80%|████████ | 4/5 [00:02<00:00, 1.99it/s]
100%|██████████| 5/5 [00:02<00:00, 2.35it/s]
100%|██████████| 5/5 [00:02<00:00, 2.33it/s]
0%| | 0/5 [00:00<?, ?it/s]
20%|██ | 1/5 [00:00<00:01, 2.03it/s]
100%|██████████| 5/5 [00:02<00:00, 2.38it/s]
100%|██████████| 5/5 [00:02<00:00, 2.36it/s]
0%| | 0/5 [00:00<?, ?it/s]
100%|██████████| 5/5 [00:02<00:00, 2.00it/s]
100%|██████████| 5/5 [00:02<00:00, 2.00it/s]
20%|██ | 2/10 [00:02<00:09, 1.22s/it]
0%| | 0/5 [00:00<?, ?it/s]
20%|██ | 1/5 [00:00<00:01, 2.38it/s]
20%|██ | 1/5 [00:00<00:01, 2.12it/s]
40%|████ | 2/5 [00:01<00:01, 1.97it/s]
20%|██ | 1/5 [00:00<00:01, 2.34it/s]
40%|████ | 2/5 [00:00<00:01, 2.41it/s]
40%|████ | 2/5 [00:00<00:01, 2.11it/s]
60%|██████ | 3/5 [00:01<00:01, 1.97it/s]
40%|████ | 2/5 [00:00<00:01, 2.31it/s]
60%|██████ | 3/5 [00:01<00:00, 2.43it/s]
60%|██████ | 3/5 [00:01<00:00, 2.09it/s]
80%|████████ | 4/5 [00:01<00:00, 2.45it/s]
60%|██████ | 3/5 [00:01<00:00, 2.27it/s]
80%|████████ | 4/5 [00:02<00:00, 1.96it/s]
100%|██████████| 5/5 [00:02<00:00, 2.43it/s]
100%|██████████| 5/5 [00:02<00:00, 2.42it/s]
80%|████████ | 4/5 [00:01<00:00, 2.05it/s]
80%|████████ | 4/5 [00:01<00:00, 2.23it/s]
100%|██████████| 5/5 [00:02<00:00, 1.99it/s]
100%|██████████| 5/5 [00:02<00:00, 1.98it/s]
100%|██████████| 5/5 [00:02<00:00, 2.60it/s]
100%|██████████| 5/5 [00:02<00:00, 2.44it/s]
100%|██████████| 5/5 [00:02<00:00, 2.36it/s]
100%|██████████| 5/5 [00:02<00:00, 2.23it/s]
30%|███ | 3/10 [00:04<00:11, 1.59s/it]
--------------------------------------------------------------------------- RemoteTraceback Traceback (most recent call last) RemoteTraceback: Traceback (most recent call last): File "/home/runner/work/modelbase2/modelbase2/.venv/lib/python3.13/site-packages/pebble/common.py", line 183, in process_execute return Result(SUCCESS, function(*args, **kwargs)) ~~~~~~~~^^^^^^^^^^^^^^^^^ File "/home/runner/work/modelbase2/modelbase2/src/modelbase2/parallel.py", line 83, in _load_or_run res = fn(v) File "/home/runner/work/modelbase2/modelbase2/src/modelbase2/scan.py", line 64, in _update_parameters_and return fn(model) File "/home/runner/work/modelbase2/modelbase2/src/modelbase2/mca.py", line 262, in response_coefficients res = parallelise( partial( ...<11 lines>... max_workers=max_workers, ) File "/home/runner/work/modelbase2/modelbase2/src/modelbase2/parallel.py", line 163, in parallelise results = dict( tqdm( ...<4 lines>... ) # type: ignore ) # type: ignore File "/home/runner/work/modelbase2/modelbase2/.venv/lib/python3.13/site-packages/tqdm/std.py", line 1181, in __iter__ for obj in iterable: ^^^^^^^^ File "/home/runner/work/modelbase2/modelbase2/src/modelbase2/parallel.py", line 83, in _load_or_run res = fn(v) File "/home/runner/work/modelbase2/modelbase2/src/modelbase2/mca.py", line 195, in _response_coefficient_worker upper = _steady_state_worker( model, y0=y0, rel_norm=rel_norm, ) File "/home/runner/work/modelbase2/modelbase2/src/modelbase2/scan.py", line 328, in _steady_state_worker c, v = unwrap( ~~~~~~^ Simulator(model, y0=y0) ^^^^^^^^^^^^^^^^^^^^^^^ .simulate_to_steady_state(rel_norm=rel_norm) ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ .get_result() ^^^^^^^^^^^^^ ) ^ File "/home/runner/work/modelbase2/modelbase2/src/modelbase2/types.py", line 86, in unwrap raise ValueError(msg) ValueError: Unexpected None The above exception was the direct cause of the following exception: ValueError Traceback (most recent call last) Cell In[8], line 8 2 rc = mca.response_coefficients( 3 get_lin_chain_two_circles(), 4 parameters=["vmax_1", "vmax_2", "vmax_3", "vmax_5", "vmax_6"], 5 ) 6 _ = plot.heatmap(rc.concs) ----> 8 mrc = mc.response_coefficients( 9 get_lin_chain_two_circles(), 10 parameters=["vmax_1", "vmax_2", "vmax_3", "vmax_5", "vmax_6"], 11 mc_parameters=sample( 12 { 13 "k0": LogNormal(np.log(1.0), 1.0), 14 "k4": LogNormal(np.log(0.5), 1.0), 15 }, 16 n=10, 17 ), 18 ) 20 _ = plot.violins_from_2d_idx(mrc.concs, n_cols=len(mrc.concs.columns)) File ~/work/modelbase2/modelbase2/src/modelbase2/mc.py:521, in response_coefficients(model, parameters, mc_parameters, y0, cache, normalized, displacement, disable_tqdm, max_workers, rel_norm) 478 def response_coefficients( 479 model: Model, 480 parameters: list[str], (...) 489 rel_norm: bool = False, 490 ) -> ResponseCoefficientsByPars: 491 """Calculate response coefficients using Monte Carlo analysis. 492 493 Examples: (...) 519 520 """ --> 521 res = parallelise( 522 fn=partial( 523 _update_parameters_and, 524 fn=partial( 525 mca.response_coefficients, 526 parameters=parameters, 527 y0=y0, 528 normalized=normalized, 529 displacement=displacement, 530 rel_norm=rel_norm, 531 disable_tqdm=disable_tqdm, 532 parallel=False, 533 ), 534 model=model, 535 ), 536 inputs=list(mc_parameters.iterrows()), 537 cache=cache, 538 max_workers=max_workers, 539 ) 541 crcs = {k: v.concs for k, v in res.items()} 542 frcs = {k: v.fluxes for k, v in res.items()} File ~/work/modelbase2/modelbase2/src/modelbase2/parallel.py:155, in parallelise(fn, inputs, cache, parallel, max_workers, timeout, disable_tqdm, tqdm_desc) 153 while True: 154 try: --> 155 key, value = next(it) 156 pbar.update(1) 157 results[key] = value File ~/work/modelbase2/modelbase2/.venv/lib/python3.13/site-packages/pebble/pool/base_pool.py:230, in MapResults.next(self) 226 return result.value 228 result = result.value --> 230 raise result ValueError: Unexpected None
First finish line
With that you now know most of what you will need from a day-to-day basis about monte carlo methods in modelbase2.Congratulations!
Advanced topics¶
Parameter scans¶
Vary both monte carlo parameters as well as systematically scan for other parameters
mcss = mc.scan_steady_state(
get_linear_chain_2v(),
parameters=pd.DataFrame({"k1": np.linspace(0, 1, 3)}),
mc_parameters=sample(
{
"k2": Uniform(1.0, 1.3),
"k3": LogNormal(mean=1.0, sigma=0.2),
},
n=10,
),
)
plot.violins_from_2d_idx(mcss.concs)
plt.show()
0%| | 0/10 [00:00<?, ?it/s]
0%| | 0/3 [00:00<?, ?it/s]
0%| | 0/3 [00:00<?, ?it/s]
0%| | 0/3 [00:00<?, ?it/s]
0%| | 0/3 [00:00<?, ?it/s]
100%|██████████| 3/3 [00:00<00:00, 28.88it/s]
100%|██████████| 3/3 [00:00<00:00, 29.69it/s]
100%|██████████| 3/3 [00:00<00:00, 27.26it/s]
100%|██████████| 3/3 [00:00<00:00, 28.05it/s]
100%|██████████| 3/3 [00:00<00:00, 27.00it/s]
100%|██████████| 3/3 [00:00<00:00, 24.03it/s]
0%| | 0/3 [00:00<?, ?it/s]
10%|█ | 1/10 [00:00<00:01, 5.77it/s]
100%|██████████| 3/3 [00:00<00:00, 26.41it/s]
0%| | 0/3 [00:00<?, ?it/s]
0%| | 0/3 [00:00<?, ?it/s]
100%|██████████| 3/3 [00:00<00:00, 23.45it/s]
0%| | 0/3 [00:00<?, ?it/s]
100%|██████████| 3/3 [00:00<00:00, 33.30it/s]
0%| | 0/3 [00:00<?, ?it/s]
100%|██████████| 3/3 [00:00<00:00, 27.45it/s]
100%|██████████| 3/3 [00:00<00:00, 25.14it/s]
100%|██████████| 3/3 [00:00<00:00, 27.35it/s]
100%|██████████| 3/3 [00:00<00:00, 24.92it/s]
100%|██████████| 3/3 [00:00<00:00, 29.26it/s]
100%|██████████| 3/3 [00:00<00:00, 27.63it/s]
0%| | 0/3 [00:00<?, ?it/s]
50%|█████ | 5/10 [00:00<00:00, 18.99it/s]
100%|██████████| 3/3 [00:00<00:00, 35.34it/s]
100%|██████████| 3/3 [00:00<00:00, 44.65it/s]
100%|██████████| 10/10 [00:00<00:00, 22.07it/s]
# FIXME: no idea how to plot this yet. Ridge plots?
# Maybe it's just a bit much :D
mcss = mc.scan_steady_state(
get_linear_chain_2v(),
parameters=mb2.cartesian_product(
{
"k1": np.linspace(0, 1, 3),
"k2": np.linspace(0, 1, 3),
}
),
mc_parameters=sample(
{
"k3": LogNormal(mean=1.0, sigma=0.2),
},
n=10,
),
)
mcss.concs.head()
0%| | 0/10 [00:00<?, ?it/s]
0%| | 0/9 [00:00<?, ?it/s]
0%| | 0/9 [00:00<?, ?it/s]
0%| | 0/9 [00:00<?, ?it/s]
0%| | 0/9 [00:00<?, ?it/s]
22%|██▏ | 2/9 [00:00<00:00, 14.82it/s]
22%|██▏ | 2/9 [00:00<00:00, 15.30it/s]
22%|██▏ | 2/9 [00:00<00:00, 15.17it/s]
22%|██▏ | 2/9 [00:00<00:00, 16.54it/s]
33%|███▎ | 3/9 [00:00<00:00, 11.99it/s]
33%|███▎ | 3/9 [00:00<00:00, 12.33it/s]
0%| | 0/9 [00:00<?, ?it/s]
0%| | 0/9 [00:00<?, ?it/s]
33%|███▎ | 3/9 [00:00<00:00, 11.16it/s]
33%|███▎ | 3/9 [00:00<00:00, 11.85it/s]
0%| | 0/9 [00:00<?, ?it/s]
0%| | 0/9 [00:00<?, ?it/s]
22%|██▏ | 2/9 [00:00<00:00, 19.33it/s]
22%|██▏ | 2/9 [00:00<00:00, 16.69it/s]
33%|███▎ | 3/9 [00:00<00:00, 21.42it/s]
22%|██▏ | 2/9 [00:00<00:00, 18.08it/s]
33%|███▎ | 3/9 [00:00<00:00, 14.78it/s]
0%| | 0/9 [00:00<?, ?it/s]
33%|███▎ | 3/9 [00:00<00:00, 14.00it/s]
0%| | 0/9 [00:00<?, ?it/s]
33%|███▎ | 3/9 [00:00<00:00, 12.15it/s]
33%|███▎ | 3/9 [00:00<00:00, 12.88it/s]
33%|███▎ | 3/9 [00:00<00:00, 24.82it/s]
33%|███▎ | 3/9 [00:00<00:00, 25.85it/s]
33%|███▎ | 3/9 [00:00<00:00, 19.09it/s]
33%|███▎ | 3/9 [00:00<00:00, 20.05it/s]
0%| | 0/10 [00:00<?, ?it/s]
--------------------------------------------------------------------------- RemoteTraceback Traceback (most recent call last) RemoteTraceback: Traceback (most recent call last): File "/home/runner/work/modelbase2/modelbase2/.venv/lib/python3.13/site-packages/pebble/common.py", line 183, in process_execute return Result(SUCCESS, function(*args, **kwargs)) ~~~~~~~~^^^^^^^^^^^^^^^^^ File "/home/runner/work/modelbase2/modelbase2/src/modelbase2/parallel.py", line 83, in _load_or_run res = fn(v) File "/home/runner/work/modelbase2/modelbase2/src/modelbase2/scan.py", line 64, in _update_parameters_and return fn(model) File "/home/runner/work/modelbase2/modelbase2/src/modelbase2/mc.py", line 114, in _parameter_scan_worker return scan.steady_state( ~~~~~~~~~~~~~~~~~^ model, ^^^^^^ ...<3 lines>... rel_norm=rel_norm, ^^^^^^^^^^^^^^^^^^ ) ^ File "/home/runner/work/modelbase2/modelbase2/src/modelbase2/scan.py", line 447, in steady_state res = parallelise( partial( ...<10 lines>... parallel=parallel, ) File "/home/runner/work/modelbase2/modelbase2/src/modelbase2/parallel.py", line 163, in parallelise results = dict( tqdm( ...<4 lines>... ) # type: ignore ) # type: ignore File "/home/runner/work/modelbase2/modelbase2/.venv/lib/python3.13/site-packages/tqdm/std.py", line 1181, in __iter__ for obj in iterable: ^^^^^^^^ File "/home/runner/work/modelbase2/modelbase2/src/modelbase2/parallel.py", line 83, in _load_or_run res = fn(v) File "/home/runner/work/modelbase2/modelbase2/src/modelbase2/scan.py", line 64, in _update_parameters_and return fn(model) File "/home/runner/work/modelbase2/modelbase2/src/modelbase2/scan.py", line 328, in _steady_state_worker c, v = unwrap( ~~~~~~^ Simulator(model, y0=y0) ^^^^^^^^^^^^^^^^^^^^^^^ .simulate_to_steady_state(rel_norm=rel_norm) ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ .get_result() ^^^^^^^^^^^^^ ) ^ File "/home/runner/work/modelbase2/modelbase2/src/modelbase2/types.py", line 86, in unwrap raise ValueError(msg) ValueError: Unexpected None The above exception was the direct cause of the following exception: ValueError Traceback (most recent call last) Cell In[10], line 4 1 # FIXME: no idea how to plot this yet. Ridge plots? 2 # Maybe it's just a bit much :D ----> 4 mcss = mc.scan_steady_state( 5 get_linear_chain_2v(), 6 parameters=mb2.cartesian_product( 7 { 8 "k1": np.linspace(0, 1, 3), 9 "k2": np.linspace(0, 1, 3), 10 } 11 ), 12 mc_parameters=sample( 13 { 14 "k3": LogNormal(mean=1.0, sigma=0.2), 15 }, 16 n=10, 17 ), 18 ) 20 mcss.concs.head() File ~/work/modelbase2/modelbase2/src/modelbase2/mc.py:325, in scan_steady_state(model, parameters, mc_parameters, y0, max_workers, cache, rel_norm, worker) 281 def scan_steady_state( 282 model: Model, 283 parameters: pd.DataFrame, (...) 290 worker: ParameterScanWorker = _parameter_scan_worker, 291 ) -> McSteadyStates: 292 """Parameter scan of mc distributed steady states. 293 294 Examples: (...) 323 324 """ --> 325 res = parallelise( 326 partial( 327 _update_parameters_and, 328 fn=partial( 329 worker, 330 parameters=parameters, 331 y0=y0, 332 rel_norm=rel_norm, 333 ), 334 model=model, 335 ), 336 inputs=list(mc_parameters.iterrows()), 337 cache=cache, 338 max_workers=max_workers, 339 ) 340 concs = {k: v.concs.T for k, v in res.items()} 341 fluxes = {k: v.fluxes.T for k, v in res.items()} File ~/work/modelbase2/modelbase2/src/modelbase2/parallel.py:155, in parallelise(fn, inputs, cache, parallel, max_workers, timeout, disable_tqdm, tqdm_desc) 153 while True: 154 try: --> 155 key, value = next(it) 156 pbar.update(1) 157 results[key] = value File ~/work/modelbase2/modelbase2/.venv/lib/python3.13/site-packages/pebble/pool/base_pool.py:230, in MapResults.next(self) 226 return result.value 228 result = result.value --> 230 raise result ValueError: Unexpected None
Custom distributions¶
If you want to create custom distributions, all you need to do is to create a class that follows the Distribution protocol, e.g. implements a sample function.
For API consistency, the sample method has to take rng argument, which can be ignored if not applicable.
from dataclasses import dataclass
from typing import TYPE_CHECKING
if TYPE_CHECKING:
from modelbase2.types import Array
@dataclass
class MyOwnDistribution:
loc: float = 0.0
scale: float = 1.0
def sample(
self,
num: int,
rng: np.random.Generator | None = None,
) -> Array:
if rng is None:
rng = np.random.default_rng()
return rng.normal(loc=self.loc, scale=self.scale, size=num)
sample({"p1": MyOwnDistribution()}, n=5)
| p1 | |
|---|---|
| 0 | 0.714775 |
| 1 | 0.861520 |
| 2 | 0.048160 |
| 3 | 1.344371 |
| 4 | 0.025791 |